

The package consists of a C++ backend, which performs the calculations, and a Python frontend, which serves as API for the user. In this application note we present a new, complete, and self-contained package, which calculates dihedral entropies on an input data set of dihedral angles. The sum of these local entropies can be considered an approximation of the total entropy in the system, i.e., the approximation that neglects all higher order terms to the entropy. (16) In contrast to our approach, these aim at calculating the total entropy of the entire system whereas our approach calculates localized entropies of the individual residues. There are other approaches for calculating the dihedral entropy, e.g., quasiharmonic calculation, (13) 2D Entropy, (14) MIST, (15) or the use of Gaussian Mixtures. It is worth mentioning that we are calculating the classical coordinate-based dihedral entropy and use a 1D approximation of the entropy. The entropy is then calculated by integrating the probability density functions (PDF) of the individual backbone dihedral angle distributions of the simulated protein.
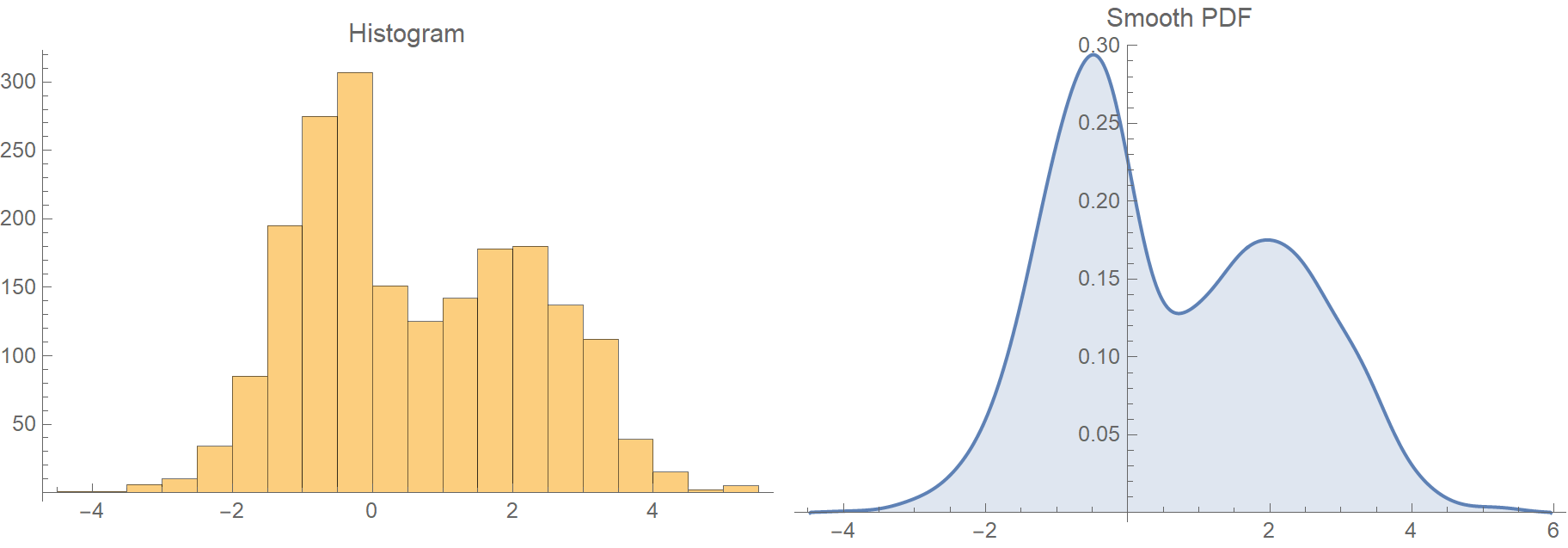
The presented approach is based on a plug-in bandwidth selection method presented by Botev et al., (12) which facilitates automatized bandwidth optimization. (10,11) A major advantage of this metric is that it is completely alignment-independent and quantifies local flexibilities directly from the fundamental thermodynamics encoded in the simulation. In this work we focus on a residuewise dihedral entropy metric, previously presented by our group. Typical analysis approaches range from interaction- or contact-based analyses, like H-bond, ionic, or native contact analyses, over structural characterizations like 1D- and 2D-RMSD, or clustering analyses, to flexibility metrics, like RMSF, or conformational entropy analyses. As the motions of biomolecules comprise large domain movements as well as delicate side-chain rearrangements, numerous analysis tools are available to characterize the captured ensembles. (3,9) One major challenge in working with MD simulations is the intractable complexity of the raw output data. Molecular dynamics (MD) simulations are a vital tool to study the conformational flexibility of biomolecules, as they capture conformational ensembles in atomistic detail with reliable state probabilities. In particular as the relevance of biopharmaceuticals steadily increases, also the thorough exploration of protein dynamics becomes ever more important. Thus, improving our fundamental understanding of these mechanisms relies on a robust characterization of conformational ensembles. (3−5) It is well established that countless physiological processes, such as biomolecular recognition, (6−8) catalytic activity, (2) or drug binding, (9) are directly linked to a biomolecule’s conformational ensemble. (1,2) Therefore, all physicochemical properties correspond to an ensemble of structures with varying probabilities and not to a single structure alone.
#Kernel density estimation free
X-Entropy is available free of charge on GitHub ( ).īiomolecules constantly fluctuate between various conformations. Further, we analyze the computational performance of this module compared to well-established python libraries that perform KDE analyses. In particular, we benchmark the performance of our module in calculating the entropy of samples drawn from a Gaussian distribution and the analytical solution thereof. In this application note, we discuss implementation and usage details and illustrate potential applications.
#Kernel density estimation full
Furthermore, the frontend allows full access to the C++ backend, so that the KDE can be used on any binnable one-dimensional input data. This makes the package very straightforward to include in any Python-based analysis workflow. We further provide a Python frontend, with predefined wrapper functions for classical coordinate-based dihedral entropy calculations, using a 1D approximation. The key feature of our approach is a Gaussian kernel density estimation (KDE) using a plug-in bandwidth selection, which is fully implemented in a C++ backend and parallelized with OpenMP. The dihedral entropy facilitates an alignment-independent measure of local protein flexibility. X-Entropy is a Python package used to calculate the entropy of a given distribution, in this case, based on the distribution of dihedral angles.


 0 kommentar(er)
0 kommentar(er)
